
Members of the Bienkowski Lab at the USC Center for Integrative Connectomics, which combines spatial transcriptomics, circuit mapping, and imaging to understand how distinct brain cell types are affected in disease.
Meet the Bienkowski Lab at the USC Center for Integrative Connectomics
Dr. Michael S. Bienkowski is an Assistant Professor of Physiology and Neuroscience and of Biomedical Engineering at the University of Southern California. He serves as Director of the USC Center for Integrative Connectomics, based within the USC Mark and Mary Stevens Neuroimaging and Informatics Institute and affiliated with the Zilkha Neurogenetic Institute at the Keck School of Medicine of USC.
The Bienkowski Lab studies how brain circuits and neuronal cell types are organized in health and disease. Leveraging a highly interdisciplinary environment, the lab integrates state-of-the-art microscopy, molecular profiling, viral tracing, and computational analysis to study both animal models and human brain tissue. This comprehensive toolkit allows the team to examine brain organization across scales—from molecular identity to three-dimensional circuit architecture.
Understanding Selective Neuronal Vulnerability in Neurodegenerative Disease
A central goal of the Center for Integrative Connectomics is to understand why specific neuronal cell types are selectively vulnerable in neurodegenerative disorders such as Alzheimer’s disease. While neurodegeneration often affects broad brain regions, disease progression frequently follows precise cellular and laminar patterns—suggesting that vulnerability is encoded at the level of cell identity, connectivity, and microenvironment.
To address this question, the Bienkowski Lab integrates:
- Viral circuit tracing to map connectivity
- Spatial transcriptomics to define molecular identity
- Three-dimensional tissue imaging to preserve anatomical context
By combining these approaches, the lab seeks to uncover the cellular and molecular mechanisms that drive disease susceptibility, with the ultimate goal of informing therapeutic strategies and improving patient outcomes.
High-Fidelity Brain Sectioning for Spatially Resolved Molecular Analysis
Accurate spatial alignment across tissue sections is essential for modern neuroscience workflows—particularly for spatial transcriptomics and multiplexed RNA imaging, where small distortions can compromise data integration. Preparing brain tissue that preserves cytoarchitecture, RNA integrity, and reproducibility is therefore a critical experimental step.
The Bienkowski Lab uses the Compresstome® vibratome from Precisionary Instruments to prepare high-quality brain tissue sections from mouse models, with a focus on delicate hippocampal regions such as CA1. The Compresstome enables the generation of uniform, well-preserved sections suitable for downstream molecular and imaging applications, including:
- Multiplexed RNA imaging
- Validation of viral tracing experiments
- High-resolution fluorescence microscopy
Consistent section thickness and minimal tissue distortion allow the lab to reliably align molecular, cellular, and anatomical data across experiments.
Enabling Discovery: The Compresstome® in Action
The Compresstome vibratome is a critical component of the Bienkowski Lab’s tissue preparation pipeline. Its ability to reproducibly section fragile hippocampal tissue while preserving native architecture has been essential for integrating spatial transcriptomic datasets with anatomical analyses.
This approach was instrumental in the lab’s recent publication in Nature Communications, where the team defined a refined laminar organization of hippocampal CA1 pyramidal neuron cell types. High-quality sectioning enabled precise mapping of molecularly distinct neuronal populations across hippocampal subregions—providing a new framework for analyzing cell-type–specific vulnerability in neurodegenerative disease models.
Research Highlight: Defining CA1 Subregions by Cell-Type Organization
In their recent study, the Bienkowski Lab demonstrated that pyramidal neurons in the CA1 hippocampal region are organized into distinct laminar subpopulations with unique molecular signatures. This refined anatomical and molecular map provides critical insight into how hippocampal circuits are structured—and why certain neuronal populations may be more susceptible to degeneration than others.
The reproducibility and tissue integrity achieved using the Compresstome vibratome were key to correlating spatial transcriptomic data with three-dimensional anatomical features, enabling conclusions that would not be possible with lower-fidelity sectioning approaches.
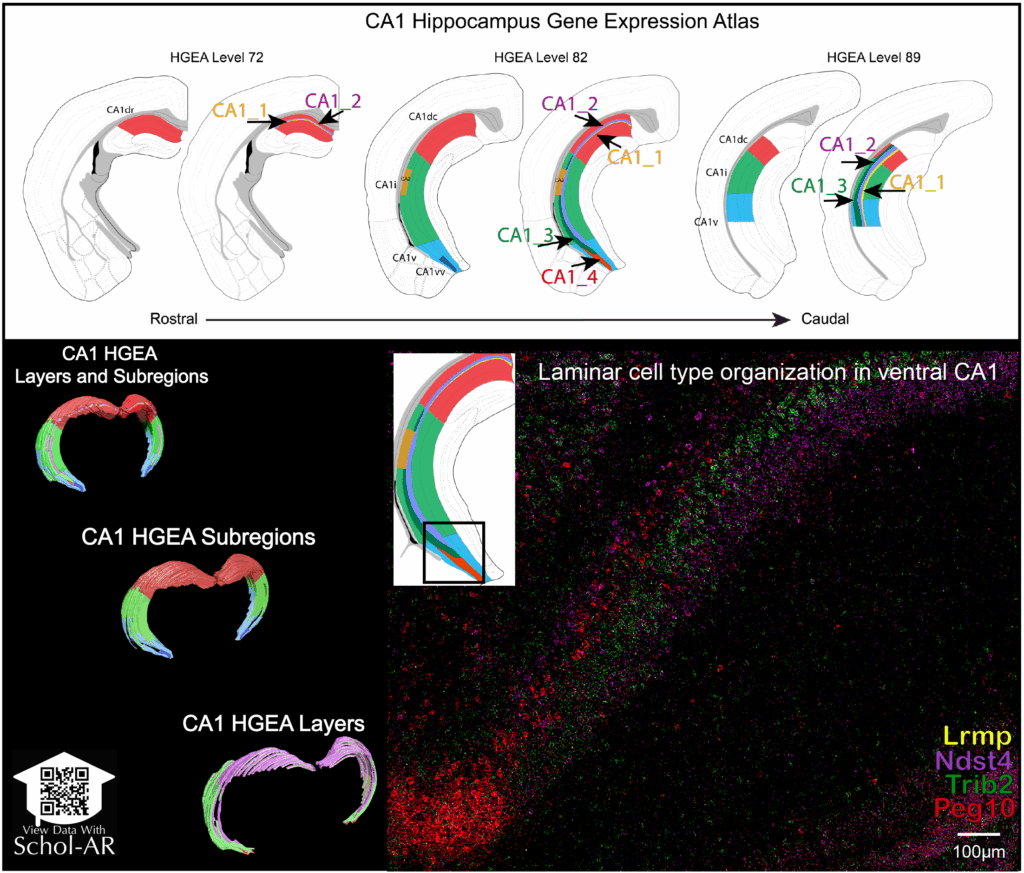
Summary of the CA1 Hippocampus Gene Expression Atlas (HGEA), illustrating laminar and subregional organization of pyramidal neuron cell types along the rostral–caudal axis. Colored domains correspond to distinct CA1 layers defined by gene expression patterns.
Looking Ahead
The Center for Integrative Connectomics continues to expand its efforts to link neuronal identity, circuit organization, and disease vulnerability. By combining precision tissue sectioning with advanced molecular and imaging technologies, the Bienkowski Lab is building a foundational framework for understanding how neurodegenerative diseases selectively target specific cells and circuits in the brain.
We are proud to support this work and look forward to seeing how these integrative approaches advance the field of neuroscience.
Connect with the Bienkowski Lab & Learn More About Precision Tissue Sectioning
Interested in learning more about the Bienkowski Lab’s research, exploring potential collaborations, or implementing high-quality brain tissue sectioning and spatial transcriptomics workflows in your own lab?
Our scientific team would be happy to connect you with the featured researchers or discuss how Precisionary Instruments can support your experimental goals—from optimizing Compresstome sectioning parameters to troubleshooting downstream imaging and molecular applications.
📬 Contact us at [email protected] to:
- Learn more about the Bienkowski Lab’s work and publications
- Explore collaboration or knowledge-sharing opportunities
- Get expert guidance on precision tissue sectioning for neuroscience research